Method validation of SYBR Green RT-PCR Assay for Identification of Strains of Pentavalent Rotavirus Vaccine
Keywords:
Validation, Identification, Rotavirus Vaccine, SYBR Green RT-PCRAbstract
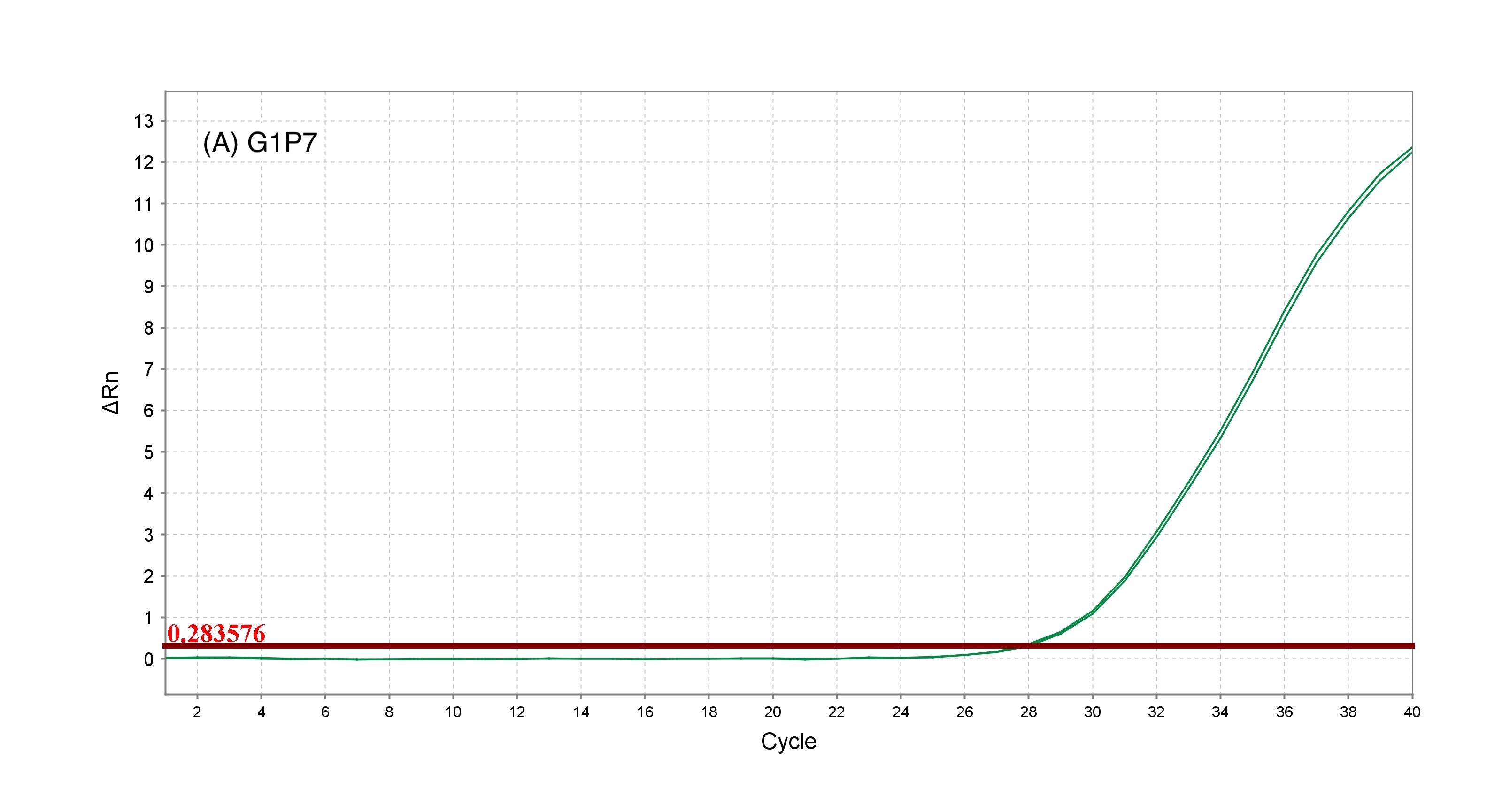
Rotavirus is a leading cause of severe gastroenteritis in infants and young children, contributing to significant global morbidity and mortality. Vaccination with rotavirus vaccines, particularly the pentavalent vaccine targeting G1P7, G2P7, G3P7, G4P7, and G6P[8] strains, provides broad protection against multiple serotypes responsible for severe disease. Accurate identification of these strains is essential for monitoring vaccine efficacy and safety. The World Health Organization (WHO) has established strict quality control guidelines to ensure the safety, efficacy, and consistency of rotavirus vaccines. This study aims to validate a SYBR Green-based reverse transcription polymerase chain reaction (SYBR Green-based RT-PCR) assay for the precise identification of strains included in the pentavalent rotavirus vaccine. The validated assay showed high specificity and reproducibility. Positive identification was confirmed for all target strains with cycle threshold (Ct) values <40.00. The assay yielded Ct values of 27.76±0.48 (G1P7), 27.76±0.74 (G2P7), 28.45±1.43 (G3P7), 28.35±0.42 (G4P7) and 30.19±1.30 (G6P[8]). Melting curve analysis confirmed specificity with single peaks at 77.03±0.03°C (G1P7), 74.50±0.07°C (G2P7), 76.42±0.21°C (G3P7), 75.58±0.17°C (G4P7), and 77.29±0.29°C (G6P[8]). Intra-assay repeatability showed %CV values of 0.14-11.47 (Ct) and 0.01-0.38 (Tm), while inter-assay reproducibility ranged from 0.68-9.51 (Ct) and 0.06-0.26 (Tm). Ruggedness/robustness testing across analysts yielded %CV 2.17-11.47 (Ct) and 0.04-0.32 (Tm). These results confirm the method’s consistency and effectively identified rotavirus strains in vaccines, supporting reliable vaccine quality control and surveillance efforts.
Downloads
References
ADAMS WR, KRAFT LM. Epizootic diarrhea of infant mice: indentification of the etiologic agent. Science. 1963;141(3578):359-360. doi:10.1126/science.141.3578.359.
Bishop RF, Davidson GP, Holmes IH, Ruck BJ. Virus particles in epithelial cells of duodenal mucosa from children with acute non-bacterial gastroenteritis. Lancet. 1973;2(7841):1281-1283. doi:10.1016/s0140-6736(73)92867-5
Omatola CA, Olaniran AO. Rotaviruses: From Pathogenesis to Disease Control-A Critical Review. Viruses. 2022;14(5):875. Published 2022 Apr 22. doi:10.3390/v14050875
Du Y, Chen C, Zhang X, et al. Global burden and trends of rotavirus infection-associated deaths from 1990 to 2019: an observational trend study. Virol J. 2022;19(1):166. Published 2022 Oct 20. doi:10.1186/s12985-022-01898-9
Gómez-Rial J, Rivero-Calle I, Salas A, et al.Rotavirus and autoimmunity. J Infect. 2020;81(2):183-189. doi:10.1016/j.jinf.2020.04.041.
Surendran S. Rotavirus infection: molecular changes and pathophysiology. EXCLI J. 2008;7:154. Doi: 0.17877/DE290R-14555
Meeting of the Strategic Advisory Group of Experts on immunization, October 2009: conclusions and recommendations. Wkly Epidemiol Rec. 2009;84(50):517-532.
World Health Organization. Recommendations to Assure the Quality, Safety and Efficacy of Rotavirus Vaccines (replacement of Annex 3 of WHO Technical Report Series, No. 941). TRS. 2007.
Ranheim T, Mathis PK, Joelsson DB, et al. Development and application of a quantitative RT-PCR potency assay for a pentavalent rotavirus vaccine (RotaTeq). J Virol Methods. 2006;131(2):193-201. doi:10.1016/j.jviromet.2005.08.013
Broeders S, Huber I, Grohmann L, et al. Guidelines for validation of qualitative real-time PCR methods. Trends Food Sci Technol. 2014;37(2):115-126. doi:10.1016/j.tifs.2014.03.008
International Organization for Standardization. ISO 20395:2019—Biotechnology: Requirements for Evaluating the Performance of Quantification Methods for Nucleic Acid Target Sequences—qPCR and dPCR. Geneva, Switzerland: ISO; 2019.
Borman P, Elder D. Q2(R1) validation of analytical procedures: text and methodology. In: Teasdale A, Elder D, Nims RW, eds. ICH Quality Guidelines. 1st ed. Hoboken, NJ: Wiley; 2017:127-166. doi:10.1002/9781118971147.ch5.
European Commission; Joint Research Centre; Institute for Health and Consumer Protection. Definition of Minimum Performance Requirements for Analytical Methods of GMO Testing: European Network of GMO Laboratories (ENGL). Luxembourg: Publications Office of the European Union; 2008. doi:10.2788/65827.
Kubista M, Andrade JM, Bengtsson M, et al. The real-time polymerase chain reaction. Mol Aspects Med. 2006;27(2-3):95-125. doi:10.1016/j.mam.2005.12.007
Mackay IM, Arden KE, Nitsche A. Real-time PCR in virology. Nucleic Acids Res. 2002;30(6):1292-1305. doi:10.1093/nar/30.6.1292
Wang Y, Liu Y, Bao H, et al. Application of the cell-based RT-qPCR assay (C-QPA) for potency detection of the novel trivalent rotavirus vaccine in China. J Clin Lab Anal. 2023;37(23-24):e24989. doi:10.1002/jcla.24989
Gautam R, Esona MD, Mijatovic-Rustempasic S, Tam KI, Gentsch JR, Bowen MD. Real-time RT-PCR assays to differentiate wild-type group A rotavirus strains from Rotarix® and RotaTeq® vaccine strains in stool samples. Hum Vaccin Immunother. 2014;10(3):767-777. doi:10.4161/hv.27388.
Tajadini M, Panjehpour M, Javanmard SH. Comparison of SYBR Green and TaqMan methods in quantitative real-time polymerase chain reaction analysis of four adenosine receptor subtypes. Adv Biomed Res. 2014;3:85. Published 2014 Feb 28. doi:10.4103/2277-9175.127998
Rehm HL, Bale SJ, Bayrak-Toydemir P, et al. ACMG clinical laboratory standards for next-generation sequencing. Genet Med. 2013;15(9):733-747. doi:10.1038/gim.2013.92
Purcell RV, Pearson J, Frizelle FA, Keenan JI. Comparison of standard, quantitative and digital PCR in the detection of enterotoxigenic Bacteroides fragilis. Sci Rep. 2016;6:34554. Published 2016 Sep 30. doi:10.1038/srep34554.
Carossino M, Barrandeguy ME, Erol E, Li Y, Balasuriya UBR. Development and evaluation of a one-step multiplex real-time TaqMan® RT-qPCR assay for the detection and genotyping of equine G3 and G14 rotaviruses in fecal samples. Virol J. 2019;16(1):49. Published 2019 Apr 25. doi:10.1186/s12985-019-1149-1
Hemming M, Vesikari T. Detection of rotateq vaccine-derived, double-reassortant rotavirus in a 7-year-old child with acute gastroenteritis. Pediatr Infect Dis J. 2014;33(6):655-656. doi:10.1097/INF.0000000000000221
Higashimoto Y, Ihira M, Miyazaki Y, et al. Monitoring Shedding of Five Genotypes of RotaTeq Vaccine Viruses by Genotype-Specific Real-Time Reverse Transcription-PCR Assays. J Clin Microbiol. 2018;56(6):e00035-18. Published 2018 May 25. doi:10.1128/JCM.00035-18
Downloads
Published
How to Cite
Issue
Section
License
Copyright (c) 2025 Chulabhorn Royal Academy

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.
Copyright and Disclaimer
Articles published in this journal are the copyright of Chulabhorn Royal Academy.
The opinions expressed in each article are those of the individual authors and do not necessarily reflect the views of Chulabhorn Royal Academy or any other faculty members of the Academy. The authors are fully responsible for all content in their respective articles. In the event of any errors or inaccuracies, the responsibility lies solely with the individual authors.


