SARS-CoV-2 : PUB and Boxing stadium in Bangkok, Thailand
Keywords:
SARS-CoV-2, COVID-19, Phylogenetic analysisAbstract
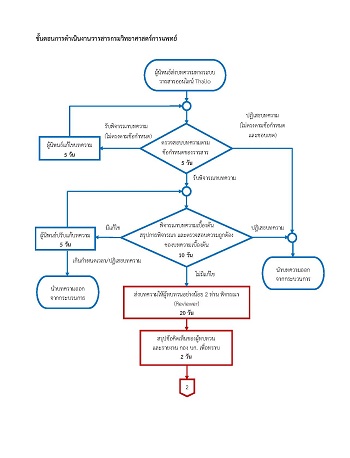
The emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in Thailand was firstly confirmed on January 8, 2020 from the patient travelling from China, and the widespread infection of COVID-19, originally from a boxing stadium and pubs in Bangkok, has rapidly occurred in March. This study was to investigate the entire genome of SARS-CoV-2 from the two groups of patients with Thai nationality. Phylogenetic analysis was performed based on whole genome sequences of isolated SARS-CoV-2 derived from clinical samples of patients. Viral genotyping for both groups revealed that the virus infection was caused by clade S with a sister lineage to the strains from South Korea and Hong Kong and a basal lineage to the strains from Spain and Mexico. Our findings showed differences from clade L that was isolated from the Chinese patient during the initial COVID-19 outbreak in Thailand. It suggested that the genetic evolution of SARS-CoV-2 in Thailand was different from that in the epicenter of the outbreak in China.
References
World Health Organization. Naming the coronavirus disease (COVID-19) and the virus that causes it. [online]. 2020; [cited 2020 Mar 1]; [1 screen]. Available from: URL: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/naming-thecoronavirus-disease-(covid-2019)-and-the-virus-that-causes-it.
Okada P, Buathong R, Phuygun S, Thanadachakul T, Parnmen S, Wongboot W, el al. Early transmission patterns of coronavirus disease 2019 (COVID-19) in travellers from Wuhan to Thailand, January 2020. Euro Surveill 2020; 25(8): 2000097. (5 pages).
Tu YF, Chien CS, Yarmishyn AA, Lin YY, Luo YH, Lin YT, et al. A review of SARS-CoV-2 and the ongoing clinical trials. Int J Mol Sci 2020; 21(7): 2657. (19 pages).
มาลินี จิตตกานต์พิชย์. ปฐมบท COVID-19. ว กรมวิทย พ. [วารสารออนไลน์]. 2563; [สืบค้น 10 เมษายน 2563]; 62(1): [5 หน้า]. เข้าถึงได้จาก: URL: https://drive.google.com/file/d/1qcVr-2ekz0P-utd-D8EPqL5OjAdMPTPin/view.
Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 2020; 395(10224): 565-74.
Marra MA, Jones SJM, Astell CR, Holt RA, Brooks-Wilson A, Butterfield YSN, et. al. The genome sequence of the SARS-associated coronavirus. Science 2003; 300(5624): 1399-404.
Phan T. Genetic diversity and evolution of SARS-CoV-2. Infect Genet Evol 2020; 81: 104260. (3 pages).
GISAID. Clade and lineage nomenclature. [online]. 2020; [cited 2020 Mar 1]; [1 screen]. Available from: URL: https://www.gisaid.org/references/statements-clarifications/clade-and-lineagenomenclature-aids-in-genomic-epidemiology-of-active-hcov-19-viruses/
Miller MA, Pfeiffer W, Schwartz T. Creating the CIPRES science gateway for inference of large phylogenetic trees. [online]. 2010; [cited 2020 Apr 10]; [7 screens]. Available from: URL: https://www.phylo.org/sub_sections/portal/sc2010_paper.pdf.
Cleemput S, Dumon W, Fonseca V, Karim WA, Giovanetti M, Alcantara LC, et al. Genome detective coronavirus typing tool for rapid identification and characterization of novel coronavirus genomes. Bioinformatics 2020; 36(11): 3552-5.
Maurier F, Beury D, Fléchon L, Varré JS, Touzet H, Goffard A, et al. A complete protocol for whole-genome sequencing of virus from clinical samples: application to coronavirus OC43. Virology 2019; 531: 141-8.
Bartolini B, Rueca M, Gruber C, Messina F, Carletti F, Giombini E, et al. SARS-CoV-2 phylogenetic analysis, Lazio Region, Italy, February-March 2020. Emerg Infect Dis 2020; 26(8): 1842-5.
Mercatelli D, Giorgi FM. Geographic and genomic distribution of SARS-CoV-2 mutations. Front Microbiol 2020; 11: 1800. (13 pages).
Shah M, Ahmad B, Choi S, Woo HG. Sequence variation of SARS-CoV-2 spike protein may facilitate stronger interaction with ACE2 promoting high infectivity. Research Square. [online]. 2020; [cited 2020 Apr 10]; [21 screens]. Available from: URL: https://assets.researchsquare.com/files/rs-16932/v1/manuscript.pdf.
Brufsky A. Distinct viral clades of SARS-CoV-2: implications for modeling of viral spread. J Med Virol 2020; 92: 1386-90.
Oran DP, Topol EJ. Prevalence of asymptomatic SARS-CoV-2 infection a narrative review. Ann Intern Med 2020; 173(5): 362-7.